Super-enhancers delineate disease-associated regulatory nodes in T cells
Nature. 2015 Feb 16. doi: 10.1038/nature14154. [Epub ahead of print]
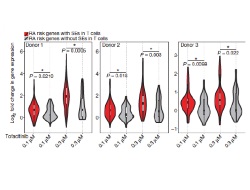
By delineating SEs, the investigators showed that these SE regions seem to occur around cytokines and their receptors in T cells. In relation to RA, 24% of RA related single-nucleotide polymorphisms (SNPs; i.e. areas of the genome that are associated with the likelihood of getting RA, or of progressing once the disease is onset) fall within SEs of T cells. Additionally, the data revealed that genes with RA SNPs that had SEs were more vulnerable to treatment with the Jak/STAT inhibitor tofacitinib compared to genes without SEs.
Overall, this study provides a systematic approach by which the SE map of disease relevant cell types can be integrated with wider analyses of the human genome to uncover drug target genes (since the effect of a drug is likely to be stronger at SE genes).